

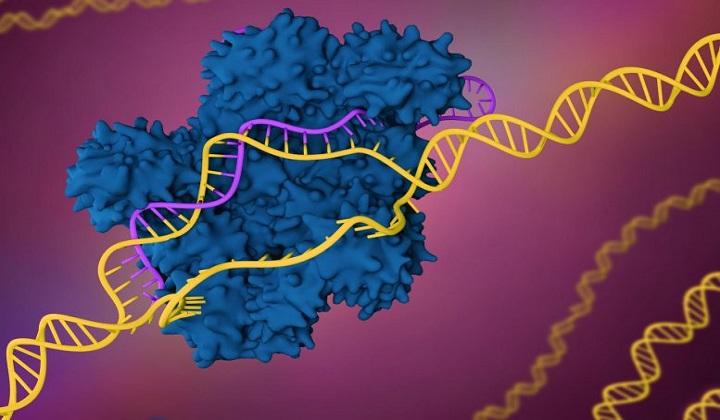
Amidst rising hopes for using CRISPR gene editing tools to repair deadly mutations linked to conditions like cystic fibrosis and sickle cell disease, a new study published today (December 6, 2019) in the Nature journal Communications Biology describes a new innovation that could accelerate this work by rapidly revealing unintended and potentially harmful changes introduced by a gene editing process.
“We’ve developed a new process for rapidly screening all of the edits made by CRISPR, and it shows there may be many more unintended changes to DNA around the site of a CRISPR repair than previously thought,” said Eric Kmiec, Ph.D., director of ChristianaCare’s Gene Editing Institute and the principal author of the study.
The study describes a new tool developed at the Gene Editing Institute that in just 48 hours can identify “multiple outcomes of CRISPR-directed gene editing,” a process that typically required up to two months of costly and complicated DNA analysis.
Kmiec cautioned that the unintended changes revealed by their work involve “subtle mutations” to DNA around the immediate site of the genome targeted for repair. That’s very different, he said, from the hotly debated concern about the risk of CRISPR causing “off-target” mutations by drifting far afield from the intended site and making random cuts across the genome.
“It’s important to note that in all instances we were still seeing CRISPR achieve a fantastic level of successful repairs that would have been unimaginable even five years ago,” added lead author Brett Sansbury. “But we saw a lot of other changes to DNA near the site of the repair that need to be better understood so that when we correct one problem, we’re not creating another.”
She said those changes included deletions, duplications and rearrangements of DNA code. And while the researchers believe the vast majority of these unintended edits may have no consequence for patients, it’s important to identify them and determine which ones might pose a risk. For example, Kmiec said unintended changes to DNA code—which acts like software for determining how genes function—could instruct a gene to produce a harmful protein.
According to the study, “such information forms the basis for determining risk-benefit decisions surrounding the effectiveness of genetic engineering tools to treat human disease.”
“CRISPR will probably never be perfect 100 percent of the time,” Kmiec said. “But CRISPR tools are constantly improving. And if we can achieve a 70 or 80 percent rate of precision—and reveal and understand the importance of any changes that occur alongside that repair—that brings us much closer to safely using CRISPR to treat patients. We hope our new tool can help accelerate efforts to achieve that goal.”
CRISPR stands for “clustered regularly interspaced short palindromic repeats.” It is a defense mechanism found in bacteria that can recognize and slice up the DNA of invading viruses.
Scientists have learned how to modify this mechanism so it can be directed to “edit” specific sequences of DNA code, with a focus on repairing DNA mutations that cause deadly diseases. For example, there is work under way to use CRISPR to repair the genetic mutation that produces the abnormal red blood cells in patients with sickle cell disease and the mutation that causes the damaging buildup of mucus in patients with cystic fibrosis.
But Kmiec noted that most tools for analyzing CRISPR gene edits are best suited for verifying that the repair was successful, not for revealing alterations that may occur to nearby strands of DNA. He said going further and screening for these unintended edits has required extracting and analyzing an enormous amount of DNA code from a cell, a sort of needle-in-a-haystack process that can take up to two months. And even then, it might not capture all the changes. The study warns that as a result, when researchers report success in using CRISPR to repair malfunctioning genes, they “may inadvertently underreport the collateral activity of this remarkable technology.”
Scientists at the Gene Editing Institute found a way around this problem by working with a system they have developed that performs gene edits on circular segments of DNA extracted from the cell, which are known as plasmids. The researchers found that working in a plasmid or “cell-free” system eliminated a lot of the complex biological activity within a cell that makes it hard to isolate the full array of DNA changes introduced by CRISPR.
In the study, they report that their system allowed them to “visualize the wide array of genetic modifications created through the process of CRISPR-directed gene editing in a straight-forward and simple fashion.” Also, because the tool can screen outcomes of an edit quickly and affordably, the researchers note that it frees scientists to execute and screen multiple trial edits—many more than is practical with a cell-based system. And that would allow them to identify unintended mutations that may occur at relatively rare frequencies and thus would otherwise go unnoticed.
It also allows them to test different variations of CRISPR. When scientists use CRISPR tools to edit a gene, they can employ different enzymes (such as Cas9 or Cas12a) to do the actual cutting and often include something called a DNA “template” to act as a map to identify and repair damaged code. The new study found that the rate of “precise repair”—repairs that are accomplished without introducing unintended mutations—varied considerably depending on the enzyme and template employed, ranging from a low of five percent to a high of 64 percent.
The work to develop a better way to screen for CRISPR-induced mutations is part of a broader effort at the Gene Editing Institute that is producing groundbreaking advances in editing DNA plasmids extracted from human cells. The team already has used their “cell-free” approach to engineer multiple edits simultaneously. This work has led to a collaboration with a biotech firm to develop new approaches to personalized cancer care. A tool developed by the team at the Gene Editing Institute can rapidly reproduce, in a human DNA sample, the unique and complex genetic features of an individual patient’s cancer tumor. And those samples can be used to screen multiple chemotherapies and other cancer drugs to design a treatment best suited for the individual patient.
Reference:
“Understanding the diversity of genetic outcomes from CRISPR-Cas generated homology-directed repair” by Brett M. Sansbury, Amanda M. Hewes and Eric B. Kmiec, 6 December 2019, Communications Biology.
Source:
https://scitechdaily.com/new-tool-for-rapidly-analyzing-crispr-edits-reveals-frequent-unintended-dna-changes/

Scheduled Server Maintenance and System Downtime Notice Dec 16, 2025
Celebrating CM Editorial Board Members Recognized in the Wor... Oct 10, 2025

Food Science and Engineering Now Indexed in CAS Database Aug 20, 2025
Contemporary Mathematics Achieves Significant Milestone in 2... Jun 19, 2025

Three Journals under Universal Wiser Publisher are Newly Ind... Apr 21, 2025